Introduction
There are various algorithms we can use to classify text. In Machine Learning, we often don't use any context or meaning representations to classify text. A document is often only represented as a bag of words. While this approach might seem too simplistic and not taking complex word interactions into account (you could say it's Naïve), it is usually good enough to classify a document as belonging to a particular class.
In this post, we'll be exploring 4 methods used to classify text using the bag-of-words approach, we'll see the code and math required, and then I'll leave you with improvements we could make to these techniques in order to improve accuracy.
The four algorithms we'll be using are:
- Bernoulli Naïve Bayes
- Multinomial Naïve Bayes
- Linear Discriminant Analysis (with TF-IDF pre-processing)
- Least Squares (with TF-IDF)
Naïve Bayes 1
The equation for Naive Bayes classification rule is,
The probability represents the probability of feature in training data given that we know the document belongs to a class .
We multiply the probabilities of each of the features appearing given that they belong to class i.e. and to obtain the overall probability, we multiply the result with the probability that a document belongs to the class in the set of all documents. We calculate these probabilities for every class and then compare them.
The largest probability is the predicted class.
Let's understand this through an example:
Say we have the following words: Gold, Bank, Dear, Hangout and we wanted to find out whether the email was spam or normal, then we can use Bayes to figure that out, effectively classifying it as spam (S) or not spam (N).
Let's say we calculated the following probabilities from word histograms:
Say our message is "Dear Bank", the probability that it belongs to class N is:
And, the probability that it belongs to class S is:
As 0.0319 > 0.008, argmax function will tell us that this email belongs to class N, i.e. it is a normal email.
Say now our message is "Hangout Bank Gold Gold", the probability that it belongs to class N is:
And, the probability that it belongs to class S is:
Even though we have a good idea that this should be classified as spam, the entire probability becomes zero as
We have two issues here:
- Zero probabilities
- Underflow (values so small, they are basically zero)
To address the issue of Zero Probabilities, we add some count (usually 1) to each word's frequency, thus obtaining non-zero probabilities for each feature. This is usually denoted by .
Also, as you can see, results in a very tiny number. If we had 100 features/words, this number might become too small for computer systems to store as there may not be enough precision in even float64 data type.
To address the issue of Underflow, we usually take log of all the probabilities and compare the log products. Since we are only comparing the relative magnitudes, any transformation that preserves proportionality while keeping the numbers large enough to store in float64 will work, and log is perfect for this.
Bernoulli Naïve Bayes 2
The decision rule for Bernoulli Naive Bayes is:
Multinomial Naïve Bayes 3
If you want to skip to the code, please check data pre-processing first and then the code for MultinomialNB here.
Now that we have that out of the way, let's take a look at the math.
In Multinomial Naive Bayes, we apply Naive Bayes to data that is multinomially distributed (where the data is represented by counts of different features). In text classification, this means our data uses word counts.
It simply tells us that the each of the follows a multinomial distribution.
The distribution is parametrized by vectors for each class , where is the number of features (in text classification, the size of the vocabulary) and is the probability of feature appearing in a sample belonging to class .
We can estimate the parameters :
Where is the number of time a feature appears in class in the training data and is the total count of all features for class .
Sci-kit learn also performs smoothing by adding 1 to every feature and prevents zero probabilities in calculations (denoted by which we covered in Naive Bayes section).
TF-IDF, LDA and Least Squares
These two topics involve a lot more math and I feel they deserve a separate post. Until then, please see the last section for reading materials.
If you want to skip to the code, LDA is here and Least Squares is covered here.
Dataset
The data we'll be using is the Newsgroup dataset which can be found here: http://qwone.com/~jason/20Newsgroups/
We'll be using the Matlab/Octave processed version of the data.
The code and explanation
We'll start by importing important libraries
import os
import tarfile
import requests
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt # for data visualization purposes
import seaborn as sns # for statistical data visualization
import sklearn as sk
%matplotlib inline
# Use 3 decimal places in output display
pd.set_option("display.precision", 3)
# Don't wrap repr(DataFrame) across additional lines
pd.set_option("display.expand_frame_repr", False)
# Set max rows displayed in output to 25
pd.set_option("display.max_rows", 25)Data Pre-processing
Let's download the newgroup data and extract it.
NEWSGROUP_DATASET_URL = "http://qwone.com/~jason/20Newsgroups/20news-bydate-matlab.tgz"
gzip_file_name = NEWSGROUP_DATASET_URL.rsplit('/', 1)[1]
NEWSGROUP_FOLDER = "20news-bydate"
NEWSGROUP_GZIP_PATH = os.path.join(".", gzip_file_name)
print(NEWSGROUP_GZIP_PATH)
# downloading the file
if not os.path.exists(NEWSGROUP_GZIP_PATH):
req = requests.get(NEWSGROUP_DATASET_URL, allow_redirects=True)
# Writing the file to the local file system
with open(NEWSGROUP_GZIP_PATH, 'wb') as output_file:
output_file.write(req.content)
print("Downloaded zip file")
else:
print("Zip file already present")
# open file
if not os.path.exists(NEWSGROUP_FOLDER):
with tarfile.open(NEWSGROUP_GZIP_PATH) as f:
# extracting file
f.extractall('.')
print("De-compressed Gzip file")
else:
print("Un-compressed folder already present")Now, we can read and process the data.
DATA_PATH = "./20news-bydate/matlab/"
TRAIN_DATA_FILE = "train.data"
TRAIN_LABEL_FILE = "train.label"
TRAIN_MAP_FILE = "train.map"
TEST_DATA_FILE = "test.data"
TEST_LABEL_FILE = "test.label"
TEST_MAP_FILE = "test.map"
train_data_path = os.path.join(DATA_PATH, TRAIN_DATA_FILE)
test_data_path = os.path.join(DATA_PATH, TEST_DATA_FILE)
train_label_path = os.path.join(DATA_PATH, TRAIN_LABEL_FILE)
test_label_path = os.path.join(DATA_PATH, TEST_LABEL_FILE)
train_map_path = os.path.join(DATA_PATH, TRAIN_MAP_FILE)
test_map_path = os.path.join(DATA_PATH, TEST_MAP_FILE)
def read_data_file(file_path):
lines = None
with open(file_path, 'r') as f:
lines = f.read()
lines = lines.split("\n")
lines = [line for line in lines if line.strip()]
return lines
def load_data(file_path):
df = pd.read_csv(file_path, names=["doc_id", "word_id", "count"], sep=' ')
return df
def load_labels(file_path):
df = pd.read_csv(file_path, names=["label_id"], sep=' ')
return df
def load_map(map_file_path):
train_map_dict = {}
train_map = read_data_file(map_file_path)
for line in train_map:
if line.strip():
k, v = line.split()
train_map_dict[k] = int(v)
return train_map_dict
train_df = load_data(train_data_path)
train_df.dropna()
test_df = load_data(test_data_path)
test_df.dropna()
train_labels = load_labels(train_label_path)
train_labels.dropna()
test_labels = load_labels(test_label_path)
test_labels.dropna()
train_map = load_map(train_map_path)
test_map = load_map(test_map_path)
labels_to_label_ids = {**train_map, **test_map}
label_ids_to_labels = dict([(v, k) for k, v in labels_to_label_ids.items()])
del train_map
del test_maptrain_df = pd.merge(train_df, train_labels, left_on="doc_id", right_index=True)
test_df = pd.merge(test_df, test_labels, left_on="doc_id", right_index=True)
train_dfSo our training and test inputs should look like this:
doc_id word_id count label_id
0 1 1 4 1
1 1 2 2 1
2 1 3 10 1
3 1 4 4 1
4 1 5 2 1
... ... ... ... ...
1467128 11268 25975 2 20
1467129 11268 27356 1 20
1467130 11268 53958 1 20
1467131 11268 53974 1 20
1467132 11268 53975 1 20
1467133 rows × 4 columnsTo simplify computation, lets use a smaller vocabulary instead of the complete vocabulary.
We'll keep only those words which have appeared more than 1000 times overall across the set of all the documents.
This could have the effect of reducing the accuracy as we'll be keeping words such as articles ("a", "an" and "the") and other regularly used fluff words which don't really mean much by themselves, and can often be removed without changing the meaning of the sentence. These words are called stopwords.
# Pruning data based on word count:
# We will keep only those words which have appeared more than 1000 times in the training data.
MIN_WORD_COUNT = 1000
grouped = train_df.groupby("word_id")
grouped = grouped.agg(total_word_count=pd.NamedAgg(column="count", aggfunc="sum"))
df = grouped.reset_index()
above_1000 = df[df["total_word_count"] > MIN_WORD_COUNT]
print(above_1000["total_word_count"].describe())
# Words with more than 1000 total appearances
final_word_list = above_1000["word_id"].unique().tolist()
N_WORDS = len(final_word_list)
words_to_idx = dict([final_word_list[i], i] for i in range(len(final_word_list)))
# Prune train df
df = train_df["word_id"].isin(final_word_list)
train_df = train_df[df]
# Prune test df
df = test_df["word_id"].isin(final_word_list)
test_df = test_df[df]
# Getting the list of all unique document ids
train_doc_ids = train_df["doc_id"].unique().tolist()
test_doc_ids = test_df["doc_id"].unique().tolist()
final_doc_id_list = train_doc_ids + test_doc_ids
final_doc_id_list.sort()
del train_doc_ids
del test_doc_ids
print(len(final_doc_id_list), final_doc_id_list[:5])18760 [1, 1, 2, 2, 3]Writing the functions for creating Bernoulli and Multinomial features:
def encode_doc_bernoulli(n_words, doc_group):
word_counts_enc_list = np.zeros(n_words, dtype=np.int32)
for row in doc_group.itertuples(index=False):
word_counts_enc_list[words_to_idx[row[1]]] = 1
return word_counts_enc_list
def encode_doc_multinomial(n_words, doc_group):
word_counts_enc_list = np.zeros(n_words, dtype=np.int32)
for row in doc_group.itertuples(index=False):
word_counts_enc_list[words_to_idx[row[1]]] = row[2]
return word_counts_enc_list
def encode_documents(df, group_col, n_words, enc_type="Bernoulli"):
docs_word_count_list = []
doc_ids = df[group_col].unique().tolist()
doc_ids.sort()
grouped = df.groupby(group_col)
if enc_type == "Bernoulli":
enc_fn = encode_doc_bernoulli
elif enc_type == "Multinomial":
enc_fn = encode_doc_multinomial
else:
enc_fn = encode_doc_bernoulli
for doc_id in doc_ids:
group = grouped.get_group(doc_id)
word_counts_enc_list = enc_fn(n_words, group)
docs_word_count_list.append(word_counts_enc_list)
return docs_word_count_list
def get_labels_for_doc_ids(df):
labels = []
doc_ids = df["doc_id"].unique().tolist()
grouped = df.groupby("doc_id")
for doc_id in doc_ids:
group = grouped.get_group(doc_id)
labels.append(group.iloc[0,3])
return labelsThe encode_documents() function will process the df and return an array of document vectors where each vector contains all of the words in our vocabulary. In the case when we want Bernoulli features, we will represent each word by a 0 if not present in the document, or a 1 if it is present.
Let's start with Bernoulli Naïve Bayes:
The BernoulliNB class of sklearn library will return a model which can train on the array of document vectors and can then be used to predict the class for a new document.
# Imports for clasification
from sklearn.naive_bayes import BernoulliNB
from sklearn import metrics
x_train = np.array(encode_documents(train_df, group_col="doc_id", n_words=N_WORDS, enc_type="Bernoulli"))
x_test = np.array(encode_documents(test_df, group_col="doc_id", n_words=N_WORDS, enc_type="Bernoulli"))
y_train = np.array(get_labels_for_doc_ids(train_df))
y_test = np.array(get_labels_for_doc_ids(test_df))
# Classification using Bernoulli Naive Bayes
bnb_model = BernoulliNB(binarize=False)
bnb_model.fit(x_train, y_train)
# Testing BernoulliNB model
y_pred = bnb_model.predict(x_test)
test_accuracy = metrics.accuracy_score(y_test, y_pred)
print("Bernoulli Naive Bayes has an accuracy of {:.2f}%".format(float(test_accuracy) * 100), "against the test set.")Bernoulli Naive Bayes has an accuracy of 28.41% against the test set.Multinomial Naive Bayes Classifier
Next, we'll explore improving the accuracy by using Multinomial Naïve Bayes:
In Multinomial Naïve Bayes, instead of using a binary representation for a word in the vector for the document, we use the word frequency.
Intuitively, if a document has certain words in larger number like "football", "player", "Barcelona", "baseball", "athlete" etc, then it's more likely to fall under the sports category/class as opposed to a document having the class politics and mentioning the word "athlete" once.
# Imports for clasification
from sklearn.naive_bayes import MultinomialNB
from sklearn import metrics
x_train = np.array(encode_documents(train_df, group_col="doc_id", n_words=N_WORDS, enc_type="Multinomial"))
x_test = np.array(encode_documents(test_df, group_col="doc_id", n_words=N_WORDS, enc_type="Multinomial"))
y_train = np.array(get_labels_for_doc_ids(train_df))
y_test = np.array(get_labels_for_doc_ids(test_df))
mn_model = MultinomialNB()
mn_model.fit(x_train, y_train)
# Testing Multinomial Naive Bayes
y_pred = mn_model.predict(x_test)
test_accuracy = metrics.accuracy_score(y_test, y_pred)
print("Multinomial Naïve Bayes has an accuracy of {:.2f}%".format(float(test_accuracy) * 100), "against the test set.")Multinomial Naïve Bayes has an accuracy of 39.02% against the test set.As we can see, using Multinomial encoding for document vector does improve the accuracy of classification.
LDA with TF-IDF
Let's see how it performs using TF-IDF and LDA.
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
x_train = np.array(encode_documents(train_df, group_col="doc_id", n_words=N_WORDS, enc_type="Multinomial"))
x_test = np.array(encode_documents(test_df, group_col="doc_id", n_words=N_WORDS, enc_type="Multinomial"))
y_train = np.array(get_labels_for_doc_ids(train_df))
y_test = np.array(get_labels_for_doc_ids(test_df))
# TF-IDF processing:
tfidf_transformer = TfidfTransformer()
# train data processing
x_train = tfidf_transformer.fit_transform(x_train).toarray()
# test data processing
x_test = tfidf_transformer.transform(x_test).toarray()
clf = LinearDiscriminantAnalysis()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_train)
test_accuracy = metrics.accuracy_score(y_train, y_pred)
print("LDA model accuracy: {:.2f}%".format(float(test_accuracy) * 100), " on training data.")LDA model accuracy: 53.73% on training data.The LDA model with TF-IDF performs better than MultinomialNB, but not by much.
Least Squares
Finally, let's check how Least Squares performs.
from sklearn.preprocessing import OneHotEncoder
from sklearn import linear_model
# Preparing train and test datasets
x_train = np.array(encode_documents(train_df, group_col="doc_id", n_words=N_WORDS, enc_type="Multinomial"))
x_test = np.array(encode_documents(test_df, group_col="doc_id", n_words=N_WORDS, enc_type="Multinomial"))
y_train = np.array(get_labels_for_doc_ids(train_df))
y_test = np.array(get_labels_for_doc_ids(test_df))
# TF-IDF processing:
tfidf_transformer = TfidfTransformer()
# train data processing
x_train = tfidf_transformer.fit_transform(x_train).toarray()
# test data processing
x_test = tfidf_transformer.transform(x_test).toarray()
# Append 1.0 to every doc vector.
one_vec = np.ones((x_train.shape[0],)).reshape(-1, 1)
x_train = np.hstack((x_train, one_vec))
one_vec = np.ones((x_test.shape[0],)).reshape(-1, 1)
x_test = np.hstack((x_test, one_vec))
print(x_train.shape, x_test.shape)
# OneHotEncode the labels for training
all_labels = list(label_ids_to_labels.keys())
all_labels.sort()
all_labels = np.array(all_labels)
onehot_encoder = OneHotEncoder()
onehot_labels = all_labels.reshape(-1, 1)
onehot_encoder.fit(onehot_labels)
y_train = onehot_encoder.transform(y_train.reshape(-1, 1)).toarray()
y_trainFor least squares, we are appending a 1.0 to every document vector. This extra column in the vector acts as the bias parameter for least squares algorithm.
We also need to one hot encode every label so that the least squares algorithm will generate a matrix of size mxn where m = number of values in document vector and n = number of classes. In our case, the parameter matrix has the dimension 293x20.
We can then find the argmax of the prediction vector and find the corresponding class for that max number's position.
If we don't onehotencode our labels, Least Squares will spit out a single number for each document as y_train will be a single number for each document.
There will be atleast two problems with this, one about how to deal with fractions, and the other to do with the fact that some classes have small numbers and other large ones.
Suppose two documents belonging to different classes have the label ids 1 and 20 respectively. The parameter that Least Squares learns will be multiplied to every value in the document vector. Thus the parameter when multiplied with two different documents having common words will cause the output to favor one label id over the other.
In order to give every class an equal opportunity regardless of their assigned label number, we use OneHot Encoding technique where we create a vector having all the classes with 1 representing the class the document belongs to and rest of the classes having 0s.
Now that we understand the processing, let's train our Least Squares classifier:
# learn the parameters
w = np.linalg.lstsq(x_train, y_train)
# multiply parameters with test data
res = np.matmul(x_test, w[0])
y_pred = np.array([all_labels[np.argmax(res[i])] for i in range(len(res))])
test_accuracy = metrics.accuracy_score(y_test, y_pred)
print("Least Squares regressor has an accuracy of {:.2f}%".format(float(test_accuracy) * 100), "against the test set.")Least Squares regressor has an accuracy of 41.51% against the test set.So far, we only checked the accuracy on the test data, so let's take a look at how we'd visualize the model's performance.
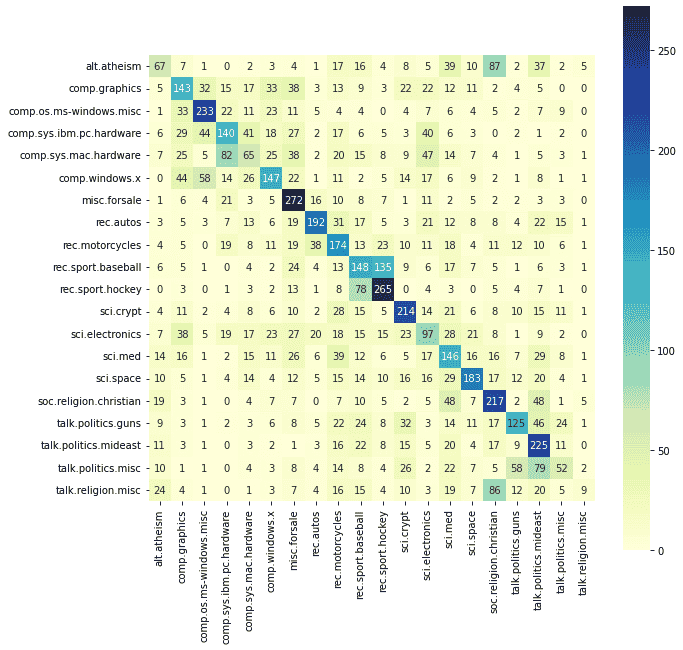
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
label_names = [label_ids_to_labels[all_labels[i]] for i in range(len(all_labels))]
cm_matrix = pd.DataFrame(data=cm, columns=label_names, index=label_names)
plt.figure(figsize = (10, 10))
sns.heatmap(cm_matrix, annot=True, square=True, fmt='d', cmap='YlGnBu')We can see that documents from the sci.electronics class are being incorrectly classified into other classes such as comp.graphics, while documents from misc.forsale are mostly being correctly identified. This makes sense as there are many classes which have an overlap with sci.electronics domain such as comp.graphics, comp.sys.mac.hardware, and sci.med as concepts in electronics are used across many domains and thus the set of words will be common across more classes.
We can also see the same happening with alt.atheism and soc.religion.christian. The vocabulary both these types of articles use will be similar and thus the model is getting "confused" between the two classes.
Visualizing data can be very powerful and can help us see the biases, outliers, etc in our data and help us decide remedial steps.
Scope for Improvements
We have hardly scratched the surface of machine learning/deep learning for text classification. There are many improvements we could make or experiments we can do to obtain a much better accuracy of classification of text documents.
Some of the improvements/experiments we can do are:
- Use a larger vocabulary (we used only the top 292 words).
- Better pre-processing (removing
stopwords, performinglemmatizationetc) - Using other algorithms such as Gradient boosted trees, Support vector machines
- Use ordering information / context by using sequence models like Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN) and their variations.
- Perform hyper-parameter tuning for models.
We could also use a much larger dataset to get better results. There's no such thing as too much data.
Conclusion
We saw four algorithms for text classification in python and implemented them and checked the results.
In the future, we can expand on this by making improvements as mentioned above.
I hope you liked this article and learned something from it. I'll be posting more articles in this space.
The Jupyter notebook is available here: https://github.com/shenoy-anurag/machine-learning/blob/main/classification-newsgroup-dataset.ipynb
References and Further Reading
- Machine Learning: A Probabalistic Approach by Kevin P. Murphy, pg 82-89; pg 103-104; pg 217-220, 2012, MIT Press, ISBN 978-0-262-01802-9 https://mitpress.mit.edu/books/machine-learning-1
- Naïve Bayes https://scikit-learn.org/stable/modules/naive_bayes.html#naive-bayes
- BernoulliNB Class Documentation https://scikit-learn.org/stable/modules/generated/sklearn.naive_bayes.BernoulliNB.html
- MultinomialNB Class Documentation https://scikit-learn.org/stable/modules/generated/sklearn.naive_bayes.MultinomialNB.html#sklearn-naive-bayes-multinomialnb
- Linear Discriminant Analysis by YANG Xiaozhou https://towardsdatascience.com/linear-discriminant-analysis-explained-f88be6c1e00b
- Least Squares https://en.wikipedia.org/wiki/Least_squares
- TF-IDF https://towardsdatascience.com/tf-idf-for-document-ranking-from-scratch-in-python-on-real-world-dataset-796d339a4089